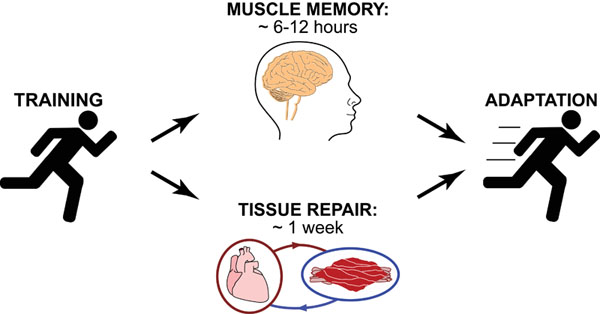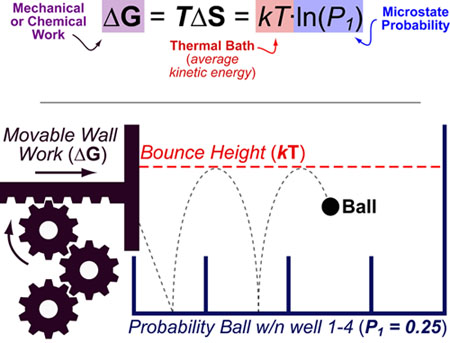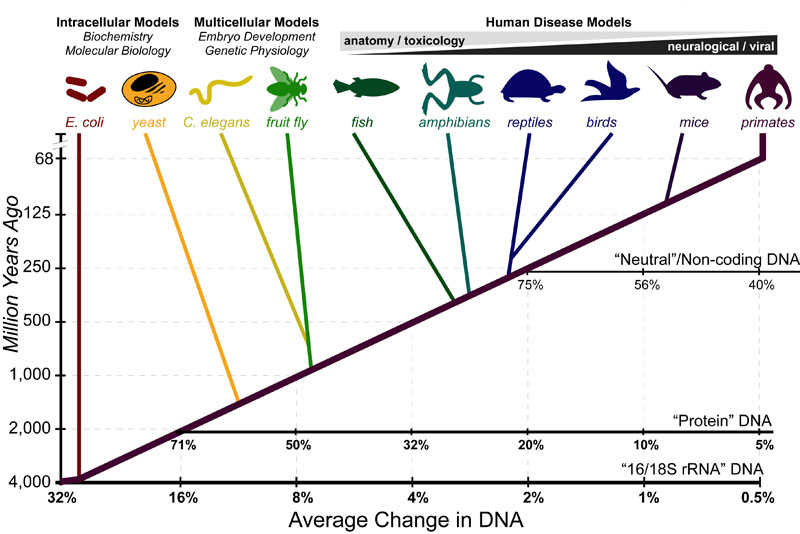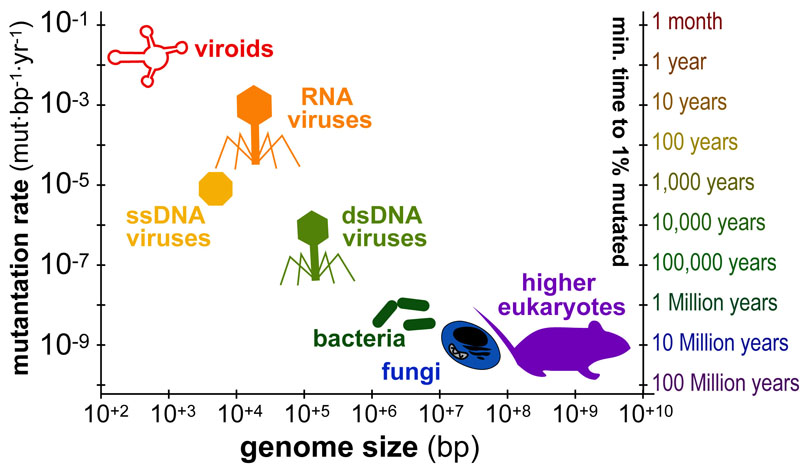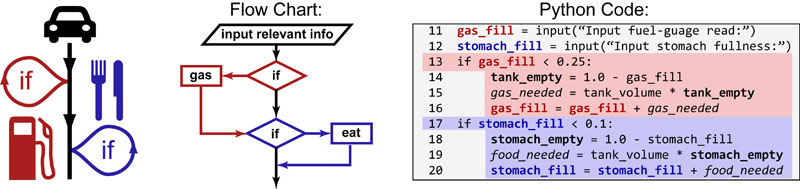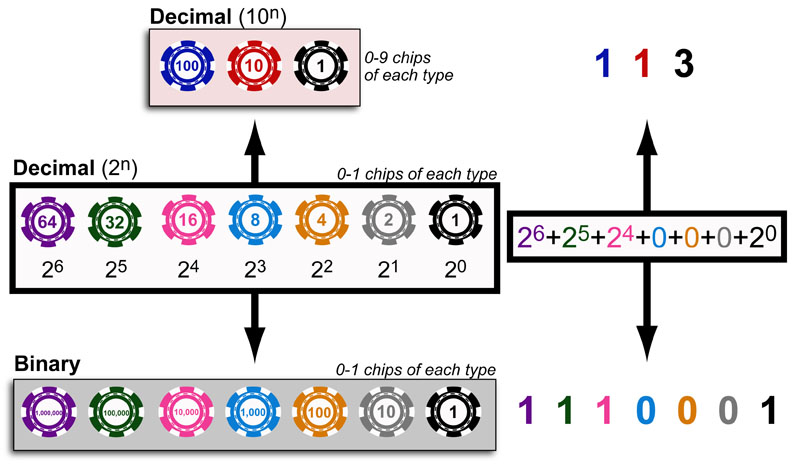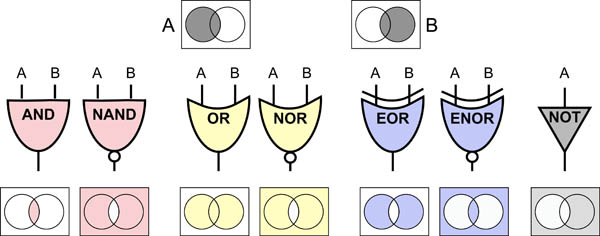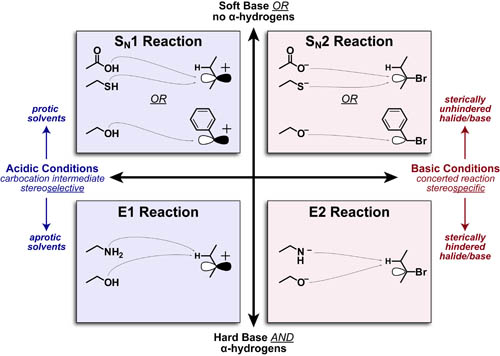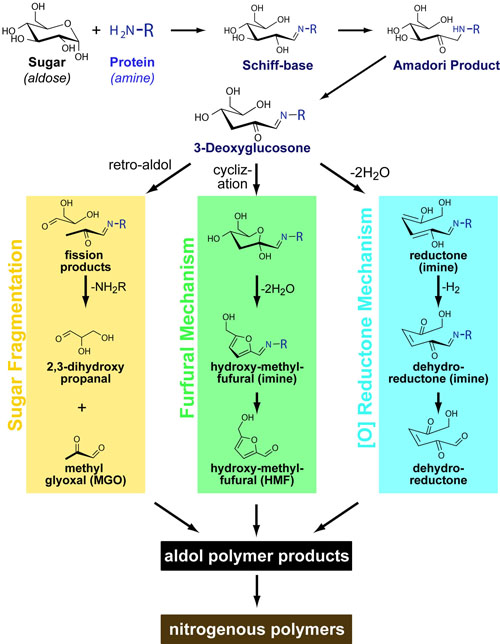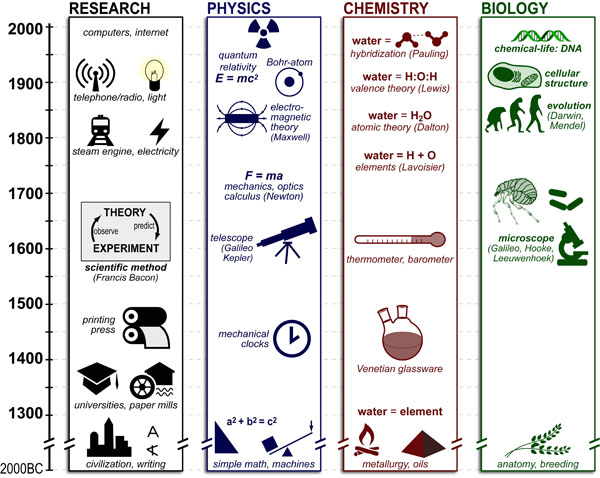
Science, as we now think of it, only really started about 400 years ago when Francis Bacon unified theory, observation, and experiment in his: “A New Method.” Before this “unified procedure,” science was a patchwork of “lucky guesses” which over-emphasized one tool or another (For example, Aristotle loved reason and hated mathematics whereas Pythagoras believed the world could only be described by pure mathematics).
In addition, quantitative experimentation and the idea of “testing a hypothesis,” only really became practically feasible with inventions of the early Renaissance. Below we give more detail on some these key milestones in the advancement of research, physics, chemistry and biology.
Continue reading →