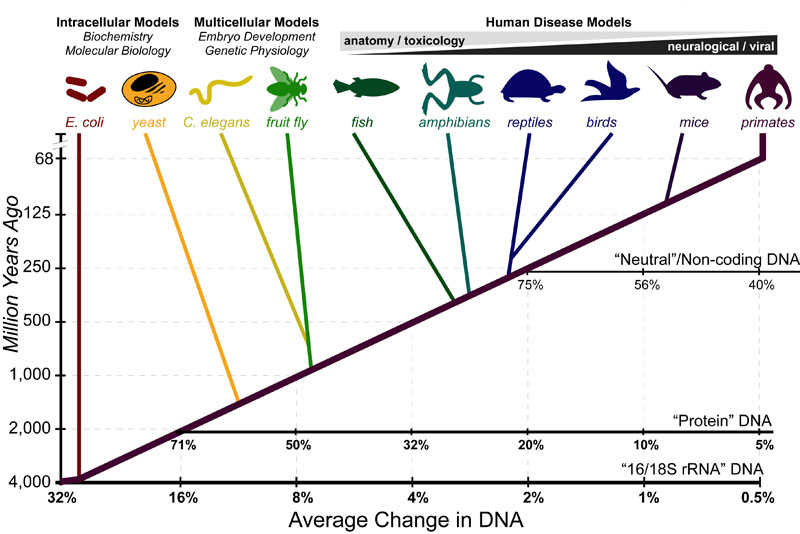
“Model organisms” are the best-studied organisms in experimental biology. For a particular research questions a specific “model organism” is chosen for its balance of: (1) ease of use and (2) “generalizability” of results. For example
- unicellular organisms(e.g. bacteria and yeast): are used to answer questions in basic biochemistry or molecular biology;
- invertebrates (e.g. worms and flys): are used to answer questions in genetics or embryonic development
- vertebrates (e.g. zebrafish to primates): are used in models of human disease (as they have requisite physiological and neurological complexity)
As, briefly discussed in our post on The Rate of Evolution, the genetic differences between these “model organism” and humans is approximately proportional to the amount of time that has elapsed since our “Last Common Ancestor” (LCA). These DNA differences have been combined with the fossil record to build the phylogenetic “Tree of Life” such as that pictured above. On average, different types of DNA evolve at different rates and can be utilized to best define different branch points of this “tree:”
- Non-coding DNA: changes the fastest and can be used to date LCA’s from ~1-100 million years ago
- Protein DNA: changes ~10x slower than non-coding DNA and can be used to date LCA’s for most (but not all) of evolutionary history
- ribosomal RNA DNA: change ~100x slower than non-coding DNA can be used to date the most ancient LCA all the way back to near the origin of life.
REFERENCES:
- Alberts, B. Molecular Biology of the Cell 5th Ed. Garland Science 2008
- Hedges, S.B.; Marin, J.; Suleski, M.; Paymer, M.; Kumar, S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 2015, 32, 835-845.
- Bier, E., McGinnis,W. Model Organisms in the Study of Development and Disease Inborn Errors of Development: the molecular bassis of clinical disorders of morphogenesis. Oxford University Press 2004
- Hedges, S.B. The Origin and Evolution of Model Organisms Nat. Rev. Gen. 2002, 3, 838-849
- Jukes,T.H.; Cantor, C.R. Evolution of Protein Molecules. New York Academic Press. 1969 21–132.
- Kimura, M. Evolutionary Rate at the Molecular Level Nature 1968, 217, 624-626
- Ochman, H.; Wilson, A.C. Evolution in Bacteria: Evidence for a Universal Substitution Rate J. Mol. Evol. 1987, 26, 74-86
- Ayala, F.J. Molecular Clock Mirages BioEssays 1999, 21, 71-75.
- Baldauf, S.L. Phyogeny for the faint of heart TRENDS in Genetics 2003, 19, 345-351.
- Kumar, S. Molecular clocks: four decades of evolution Nat. Rev. Gen. 2005, 6, 654-662.
- Scally, A.; Durbin, R. Revising the human mutation rate: implications for understanding human evolution Nat. Rev. Gen. 2012, 13, 745-753.

This work by Eugene Douglass and Chad Miller is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License.