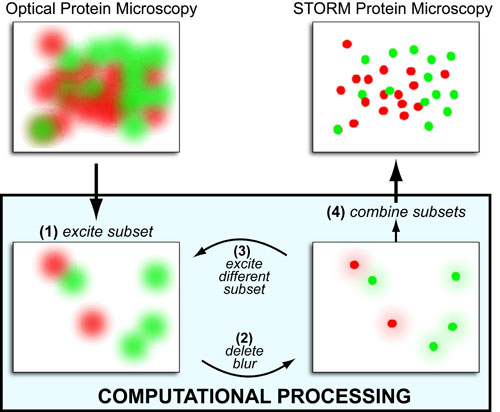
While microscopic methods with protein and atom scale resolution – that don’t break the diffraction barrier! – exist (e.g. electron microscopy, atomic force microscopy, x-ray crystallography, etc.), they tend to be more practically difficult than fluorescence-based microscopy (see posts on Fluorescent Probes and Fluorescent Antibodies). Therefore, to give fluorescence microscopy nanometer resolution, several super resolution techniques have been developed that combine clever (1) optical/photophysical and (2) computational-processing tricks to “clean up” the “blurred data.”
Given the fact that the “blurriness” (or point-spread function) of point objects (such as single proteins) can be modeled as a 2D gaussian blur a common theme in many super-resolution fluorescent microscopy techniques (i.e. single molecule localization microscopy) is to isolate subsets of those points so the full blur can be measured and computationally replaced with a single point at the blur’s center. Repeating this process iteratively, one can fully reconstruct protein-level image from a super blurry mess. Examples of such techniques include:
- Stochastic optical reconstruction microscopy (STORM)
- Photo-activated localization microscopy (PALM)
- Photo-activation localization microscopy (FPALM)
- Stimulated emission depletion (STED)
- Ground state depletion (GSD)
- Saturated structured illumination microscopy (SSIM)
- Structured illumination microscopy (SIM)
- Spatially modulated illumination (SMI)
REFERENCES:
- Small, A.; Stahlheber, S. Fluorophore localization algorithms for super-resolution microscopy Nature Methods, 2014, 11, 267-279.
- Lakowicz, J. R. in Principles of Fluorescence Spectroscopy, Springer, 2006
- Skoog, D. A., Holler, F. J. & Nieman, T. A. Principles of Instrumental Analysis (5th ed), Saunders College Publishing, 1998

This work by Eugene Douglass and Chad Miller is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License.