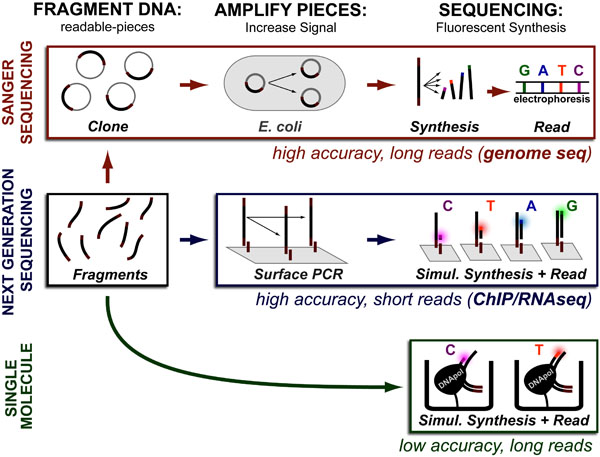
While DNA-sequencing methods are diverse and complex they can be grouped into three categories which share several common features: 1. DNA Fragmentation, 2. Fragment Amplification, 3. Sequencing via Fluorescent-Synthesis. These categories are:
Sanger-Sequencing which was invented in the 1970’s and was improved several times through the 2000’s. Sanger-Sequencing is especially good at sequencing large fragments and is often the de novo genome sequencing and variant detection.
Next Generation-Sequencing was developed by several companies in the 2000’s, is best at sequencing short fragments and is most useful when a draft genome in available. Next Generation sequencing is best at quantifying DNA copy number, gene expression levels (RNAseq), and studying chromosome dynamics (ChIPseq, ATACseq, etc.).
Finally, Single Molecule methods are the newest approaches which combine advantages and disadvantages of previous methods but are often highly portable.
REFERENCES:
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual 4th Ed., 2012, Cold Spring Harbor Laboratory Press
- Sanger, F.; Nicklen, S.; Coulson, A.R. PNAS, 1977, 74, 5463-5467
- Lohman, N.J.; Pallen, M.J. Nat. Rev. Microbio. 2015, 13, 787–794
- Pettersson, E.; Lundeberg, Ahmadian, A. Genomics, 2009, 93, 105-111
- Alberts, B. Molecular Biology of the Cell 5th Ed. Garland Science 2008
- Murphy, K. Janeway’s Immunobiology 8th Ed. Garland Science 2012

This work by Eugene Douglass and Chad Miller is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License.