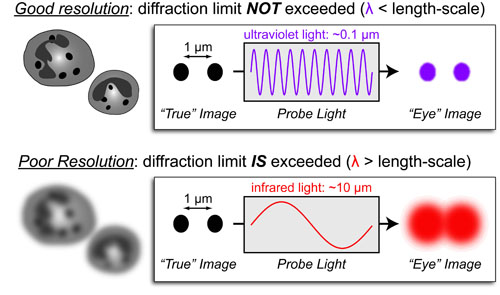
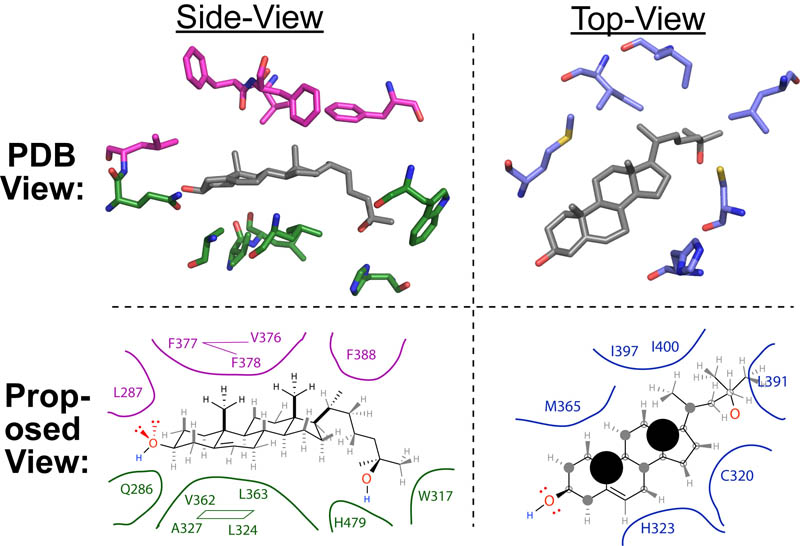
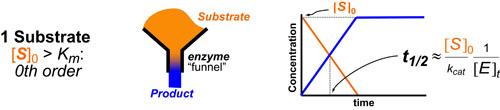
Basic light microscopy can only resolve objects that are larger than 100 nm which means that while it can visualize animal cells (~10,000nm), organelles and bacteria (~1,000nm), it cannot visualize viruses (<100nm), proteins (<10nm) or small molecules(~1nm) (see post summarizing Biological Scales). This limitation is known as the “diffraction limit” and is caused by the fact light only interacts differently with objects separated by more than one wavelength (λ). Intuitively, its helpful to think of each of these wavelengths as “a minimum pixel size” for a computer image where: infrared light (λ ~ 10.0μm) has pixels 100 times larger than ultraviolet light (λ ~ 0.1μm).
About Us
Practically Science was started by two Yale PhD students in 2012. Its goal is to make single-sheet summaries of common interdisciplinary methods, ideas, etc. Continue Reading →Please follow & like us :)
Blogroll
Site Map
-
Recent Posts