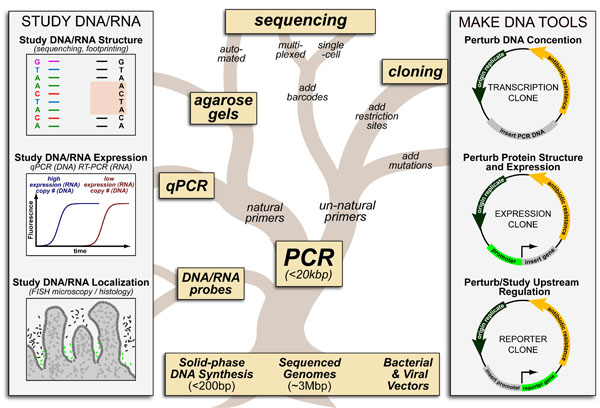
Many methods in molecular biology are simply different combinations of a handful of techniques. These combinations can be represented as a “family tree” whose “trunk”/”backbone” is PCR (polymerase chain reaction) and whose “roots”/”foundation” is built upon: (1) chemical synthesis of short oligonucleotides, (2) fully sequenced genomes and (3) vectors derived from bacteria and viruses.
DNA/RNA-BASED PROBES:
Even without PCR, these foundational technologies are sufficient to produce several short, anti-sense DNA/RNA probes including:
- Fluorescent probes: for microscopy and histology
- short interfering RNA: for gene knockdown by RNA interference
- Microarrays: for transcriptome analysis.
Due to the limits of solid-phase DNA synthesis (oligo-size < 200 kbp), PCR - which can produce oligonucleotides up to 20kbp in size - forms the backbone of most DNA based techniques described below.
QUANTITATIVE PCR FOR CONCENTRATION MEASUREMENT:
PCR makes and purifies specific DNA segments from genomic DNA, using gene-specific primer sets (). Quantitative PCR methods, uses fluorescent probes to track the PCR-amplification time course and directly measure DNA copy number (qPCR) or RNA expression level (RT-PCR). A few applications of quantitative PCR methods are listed below:
- qPCR (DNA): gene copy number, transgene copy number for GMO’s, plasmid copy number, % viral dna incorporation, bacterial/viral load per gram tissue,
- RT-PCR (DNA): mRNA expression (often a surrogate for protein expression), non-coding RNA expression, RNA viral load
AGAROSE GELS FOR STRUCTURAL QUESTIONS:
Agarose Gels are the primary means to study the structure of DNA and RNA that has been made and purified by PCR. These gels separate DNA and RNA by weight/mass and can be used to determine purity and identity or PCR products. In addition, analysis of fragmentation patterns (using nucleases, dideoxy-bases, etc.) can be used to study the primary structure (sequencing), secondary structure (DNA and RNA folding) and molecular interactions of purified DNA and RNA.
CLONING FOR SYNTHETIC DNA-BASED TOOLS:
Finally, unnatural primers can be used to add/change features of genomic DNA including the addition of: (1) mutations, (2) restriction sites and (3) barcodes. The addition of mutations and restriction sites are very important for the development of cloning tools which combine PCR-amplified DNA with a self-replicating bacterial or viral vector. Some of the most important cloning tools include
- Transcriptional clones: enable large-scale amplification of DNA within cells
- Expression clones: enable large-scale protein production and mutagenesis within cells
- Reporter clones: enable the production of cell-based fluorescent/colorimetric probes that are useful for studying signalling or high-throughput screening
REFERENCES:
- Beaucage, S.L.; Iyer, R.P. Advanced in the Synthesis of Oligonucleotides by the Phosphoramidite Approach. Tetrahedron, 1991, 48, 2223-2311.
- Molecular Cloning: Technical Guide v. 2, 2014, New England Biolabs Inc.
- Alberts, B. Molecular Biology of the Cell 5th Ed. Garland Science 2008
- Murphy, K. Janeway’s Immunobiology 8th Ed. Garland Science 2012

This work by Eugene Douglass and Chad Miller is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License.