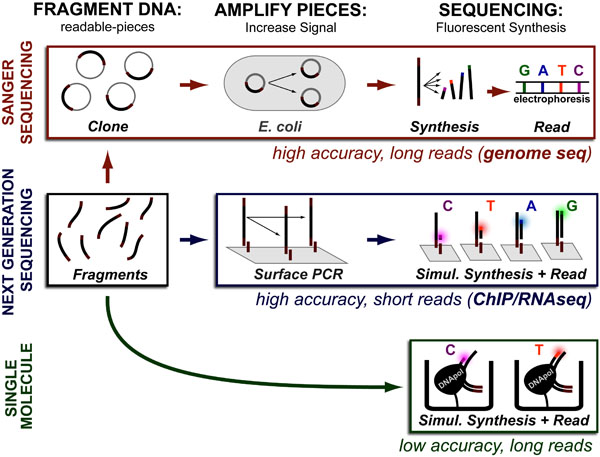
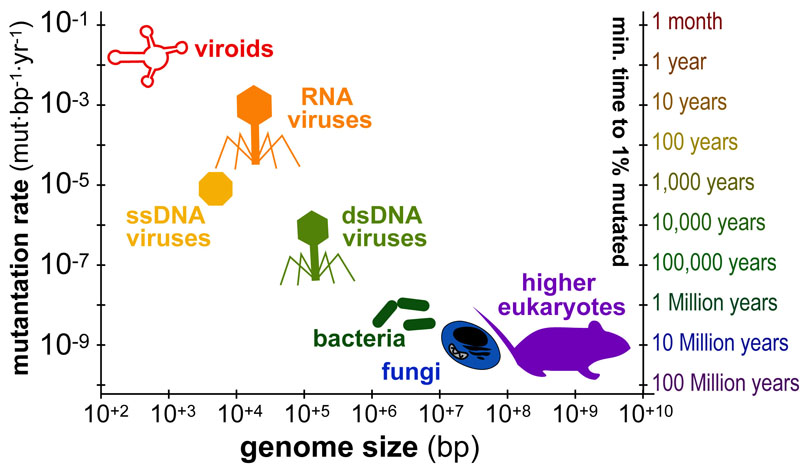
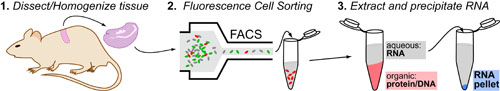
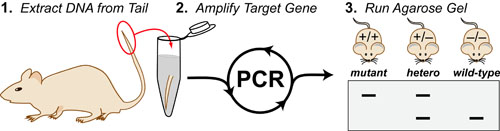
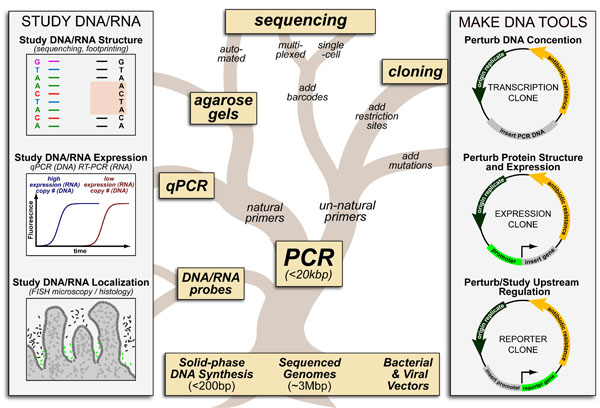
While DNA-sequencing methods are diverse and complex they can be grouped into three categories which share several common features: 1. DNA Fragmentation, 2. Fragment Amplification, 3. Sequencing via Fluorescent-Synthesis. These categories are:
Continue reading
About Us
Practically Science was started by two Yale PhD students in 2012. Its goal is to make single-sheet summaries of common interdisciplinary methods, ideas, etc. Continue Reading →Please follow & like us :)
Blogroll
Site Map
-
Recent Posts