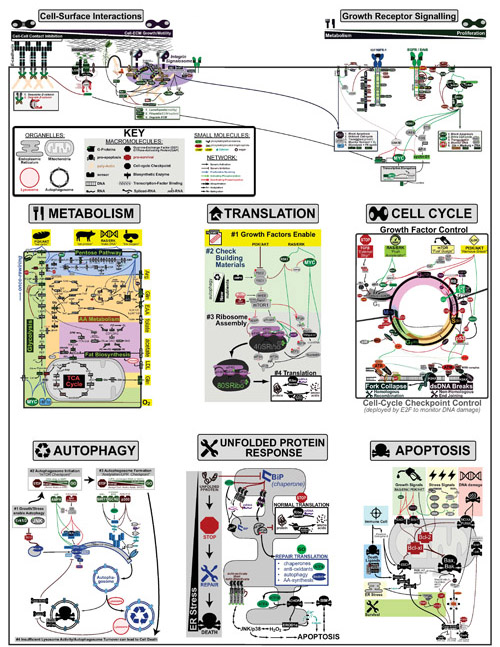
Cancer is a complex disease that is defined by at least 10 different “Hallmarks” that reflect mutation or epigenetically-driven reprogramming of normal cellular circuits. Over the past year, I have compiled a document that attempts to combine what’s known about the intra-cellular network that underlie these “Cancer Hallmarks.” This project started out as a single-page infographic but has since expanded into the 2 foot x 3 foot poster pictured above.
My goal was (and is) to create a comprehensive network map that is conceptually accessible to help me (and now others) think about the “big picture” of cancer networks. Of course, this poster is a work in progress and I will continue to update it over time. Below, I give a brief conceptual description of each module I have used to organize this “Canonical” Cancer-Network Map.
CELL-SURFACE INTERACTIONS:
This module is drawn the emphasize the competing effects that other cells and extracellular matrix(ECM) have on a cancer cell’s decisions to proliferate and move. In general, Cell-Cell Adhesion through E-cadherin are anti-proliferative, which makes sense as neighbors who are “bumping into each other” probably don’t have room to “build an expansion on their house.” Conversely, Cell-ECM Interactions through the integrins can be pro-proliferative through growth-hormone-like signalling through a multicomponent complex I call the “Integrin Signalosome.”
GROWTH RECEPTOR SIGNALLING:
This module is drawn the emphasize the different signalling emphasis of the different growth hormone receptors Insulin-like Growth Factor Receptor (IGFR) and Epidermal Growth Factor Receptor(EGFR). While both receptors are capable of signalling through the Phosphoinositide 3-kinase(PI3K) and RAS/RAF/MEK Pathways they vary in their efficiency of inducing each. For example, through the scaffold protein IRS1, IGFR is more effective at inducing PI3K signalling relative to EGFR which is more effective at inducing RAS/RAF/MEK signalling. Though these pathways are related they have distinct functional roles.
METABOLISM:
This module is drawn to further divide metabolic enzymatic “assembly lines” into 5 sub-modules:
- Glycolysis (the breakdown of sugar for energy and construction materials),
- Pentose Pathway (the use of sugar and amino acids to build DNA and maintain redox homeostasis),
- Amino Acid(AA) Metabolism (the biosynthesis and breakdown of AA for energy),
- Fat Biosynthesis (the biosynthesis of sterols, sphingomyelin(SPM), Phosphatidylethanolamine(PE), Phosphatidylcholine(PC), Phosphatidylserine(PS), Triacylglycerol(TAG), Phosphatidylinositol phosphates(PIPx)
- The TCA Cycle (the use of sugar and amino acids to extract maximal energy).
In addition, signalling inputs that can rewire metabolism are highlighted in yellow at the top and nutrient inputs are highlighted in yellow on the right hand side (EAA: Essential Amino Acids; LDL: low density lipoprotein). Genes and pathways that are upregulated in cancer have green titles (e.g. Glycolysis, Pentose Pathway, AA Metabolism, Fat Biosynthesis) while those that are downregulated in cancer have red titles (e.g. TCA cycle). Finally, if the rate-determining step (RDS) is known for a pathway it’s gene is marked by a “RDS.”
TRANSLATION:
This module is drawn to emphasize the stepwise process of global translation initiation using a building analogy where:
- First, Growth Factors approve the “building permit” of the cell
- Second, mTOR checks to make sure there is enough “building materials” (i.e. nutrients). If not, it recycles old materials via autophagy.
- Third, the ribosome (“construction equipment”) is assembled.
- Fourth, “construction” begins
CELL CYCLE:
This module is drawn to emphasize the cyclic nature of the cell cycle that is divided into 4 phases: G1 (Growth #1 ~12 hours), S (Synthesis ~6 hours), G2 (Growth #2 ~4 hours), M (Mitosis ~1 hours). The top half of the cycle is controlled by growth factors while the bottom half if controlled by “cell-cycle checkpoint machinery” which monitors DNA-damage during the process of cell-division.
I have used an automobile analogy to conceptually illustrate the roles of different growth-factor pathways in controlling the initiation of the cell cycle:
- The TGFβ pathway acts as an “emergency stop” that a cell’s community can use to tell it to stop growing
- The RAS/RAF/MEK/ERK pathway acts as the “accelerator-pedal” as its signalling flux correlates with cyclin-D that initiates the cell-cycle
- The PI3K/AKT pathway deploys a fuel guage (mTOR) and removes the break in the form of FOXO and p53 checkpoints that prevent proliferation in the context of MYC-signalling alone.
Finally, Cell-cycle Checkpoints are mostly induced by multicomponent complexes that form around double-strand DNA(dsDNA) breaks and Fork Collapses (that occur during DNA-copying). Double-Strand DNA breaks can be repaired via Non-Homologous End-joining or Homologous Recombination while Fork Collapses are usually repaired by Homologous Recombination.
AUTOPHAGY:
This module is drawn to emphasize the stepwise process of autophagy where:
- Growth or Stress signals enable Autophagy if it proves necessary
- mTOR initiates Autophagy in the context of low nutrients
- Further checks on the energy state (Acetyl-CoA, NAD+) or protein stress (Unfolded Protein Response) of the cell facilitate the formation of the Autophagosome around dysfunctional cytoplasmic compartments.
- With sufficient lysosomal activity, the dysfunctional cytoplasm material is recycled.
UNFOLDED PROTEIN RESPONSE:
This module is drawn to emphasize the stepwise process of the unfolded protein response(UPR) that occurs when the Endoplasmic Reticulum(ER) is overwhelmed by mis-folded protein
- Normal ER-localized translation is arrested
- Modified Repair translation begins
- Apoptosis is inititated if Repair fails
APOPTOSIS:
Cell-suicide, via apoptosis, is a complex process affected by diverse signals including: growth factors, stress, DNA damage and the immune system. In general, Bax and Bak are the “door” at the “gates of hell” (i.e. mitocondrial cell contents) that unleash death upon a cell. Anti-apoptotic proteins such as BCL-xl and BCL-2 can be thought of as the “lock on that door”.
Extending the door analogy to each of the signaling cues:
- Growth Factor Signals can be thought of as a “dead-bolt“, that reinforces the door protecting the cell
- DNA-damage and stress can be thought of as the “key“ that opens the door, killing the cell
- Immune cells and Death-Ligands(e.g. TNF, FASL) have their own “key“ but can also enter through a “back-door“ through direct caspase activation.
REFERENCES:
- Weinberg, R.A. The Biology of Cancer (2nd ed.), 2014, Garland Science.
- Murphy, K. Janeway’s Immunobiology 8th Ed. Garland Science 2012
- Alberts, B. Molecular Biology of the Cell 5th Ed. Garland Science 2008
- Goldberg, S. Clinical Biochemistry made ridiculously simple Medmaster Inc. 1997
- Milo, R.; Phillips, R. Cell Biology by the Numbers Garland Science 2015
- Devita, V.T.; Lawrence, T.S.; Rosenberg, S.A. Cancer: Principles and Practice of Oncology 10th Ed. Wolters Kluwer. 2015
- Berridge, M.J. Cell Cycle and Proliferation Cell Signalling Biology 2014; doi:10.1042/csb0001009
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death and Differentiation, 2011, 18, 571.
- Rubinstein, A.D.; Kimchi, A. Life in the balance – a mechanistic view of the crosstalk between autophagy and apoptosis J. Cell Sci., 2012 125, 5259.
- Galluzzi, L.; Pietrocola, F.; Levine, B.; Kroemer, G. Metabolic Control of Autophagy Cell, 2014, 159, 1263.
- Green, D.R.; Levine, B. To Be or Not to Be? How Selective Autophagy and Cell Death Govern Cell Fate. Cell, 2014, 65.
- Ouyang, L.; Shi, Z.; Zhao, S.; Wang, F.-T.; Zhou, T.T.; Liu, B.; Bao, J.-K. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis Cell Prolif., 2012, 45, 487.
- Reinhardt, H.C.; Schumacher, B. The p53 network: cellular and systemic DNA damage responses in aging and cancer Trends in Genetics, 2012, 28, 128.
- Cao, S.S.; Kaufman, R.J. Primer: Unfolded Protein Response. Current Biology, 2012, 22, R622.
- Hetz, C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond Nat. Rev. Mol. Cell Bio., 2012, 13, 89.
- Walter, P.; Ron, D. The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation Science, 2011, 334, 1081.
- Cantor, J.R.; Sabatini, D.M. Cancer Cell Metabolism: One Hallmark, Many Faces Cancer Discovery, 2012, OF1.
- Yizhak, K.; Chaneton, B.; Gottlieb, E.; Ruppin, E. Modeling cancer metabolism on a genome scale Mol. Sys. Bio., 2015, 11, 817.
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism Cell Metab., 2016, 23, 27.
- Cairns, R. A.; Harris, I.S.; Mak, T.W. Regulation of cancer cell metabolism Nat. Rev. Cancer, 2011, 11, 85.
- Shadel, G.S.; Horvath, T.L. Mitochondrial ROS Signaling in Organismal Homeostasis Cell, 2015, 163, 560.
- Jones, D.P. Radical-free biology of oxidative stress Am. J. Physiol. Cell Physiol., 2008, 295, C849.
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation Cell, 2011, 144, 646.
- Kolch, W.; Halasz, M.; Granovskaya, M.; Kholodenko, B.N. The dynamic control of signal transduction networks in cancer cells Nat. Rev. Cancer, 2015, 15, 515.
- Cully, M.; Downward, J. Snapshots: Ras Signalling Cell, 2008, 133, 1292.
- Frisch, R.; Downward, J. Snapshots: Class I PI3K Isoform Signalling Cell, 2013, 154, 940.
- Carracedo, A.; Salmena, L., Pandolfi, P.P Snapshots: PTEN Signaling Pathways Cell, 2008, 133, 550.
- MacDonald, B.T., Semenov, M.V., He, X. Snapshots: Wnt/beta-catenin Signaling Cell, 2007, 131, 1204.
- Semenov, M.V., Habas, R.; MacDonald, B.T., He, X. Snapshots: Wnt/beta-catenin Signaling Cell, 2007, 131, 1378.
- Kadowaki, T.; Ueki, K.; Yamauchi, T.; Kubota, N. Snapshots: Insulin Signaling Pathways Cell, 2012, 148, 624.
- Trempolec, N.; Dave-Coll, N.; Nebreda, A.R. Snapshots: p38 MAPK Signaling Cell, 2013, 152, 656.
- Jin, M.; Liu, X.; Klionsky, D.J. Snapshots: Selective Autophagy Cell, 2013, 152, 368.
- Wiseman, R.L. Haynes, C.M.; Ron, D. Snapshots: The Unfolded Protein Response Cell, 2010, 140, 590.
- Spizzo, R.; Nicoloso, M.S.; Croce, C.M.; Calin, G. Snapshots: MicroRNAs in Cancer Cell, 2009, 137, 586.
- Xenia, M.; Iiagan, G.; Kopan, R. Snapshots: Notch Signaling Pathway Cell, 2007, 128, 1246.
- Crabtree, G.R.; Schreiber, S.L. Snapshots: Ca2+-Calcineurin-NFAT Signaling Cell, 2009, 138, 210.
- Morgan, D.O.. Cell Cycle Regulators I Cell, 2008, 135, 764.
- Morgan, D.O.. Cell Cycle Regulators II Cell, 2008, 135, 974.
- Pope, B.D.; Aparicio, O.M.; Gilbert, D.M. Snapshots: Replication Timing Cell, 2013, 152, 1390.
- Verbrugge, I.; Johnstone, R.W.; Smyth, M.J. Snapshots: Extrinsic Apoptosis Pathways Cell, 2010, 143, 1192.
- Salvesen, G.S.; Ashkenazi, A. Snapshots: Caspases Cell, 2011, 147, 476.
- Kruse, J.-P.; Gu, W. Snapshots: p55 Post Translational Modifications Cell, 2008, 133, 930.
- Hardwick, J.M. Youle, R.J. Snapshots: BCL-2 Proteins Cell, 2009, 138, 404.
- Soulard, A.; Hall, M.H. Cell Snapshots: mTOR Signalling Cell, 2007, 129, 434.
- Laplante, M.; Sabatini, D.M. mTOR Signalling at a Glance J. Cell Sci. , 2009, 122, 3589.
- Reed, J.C.; Huang, Z. Apoptosis Pathways and Drug Targets Nature Reviews Drug Discovery/Molecular Cell Biology , 2004, http://www.nature.com/reviews/poster/apoptosis
- Conacci-Sorrell, M.; Zhurinsky, J.; BenZe’ev, A. The cadherin-catenin adhesion system in signaling and cancer, J. Clin. Invest., 2002, 109, 987.
- Cheng, Z.; Tseng, Y.; White, M.F. Insulin signaling meets mitochondria in metabolism Trends in Endocrinology and Metabolism, 2010, 21, 589.
- Hanke, S.; Mann, M. The Phosphotyrosine Interactome of the Insulin Receptor Family and Its Substrates IRS-1 and IRS-2* Mol Cell. Proteomics, 2008, 8.3, 519
- Harburger, D.S.; Calderwood, D.A. Integrin Signalling at a Glance J. Cell Sci., 2009, 122, 159.

This work by Eugene Douglass is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License.
Pingback: Haftanın posteri: Kansere giden yollar… – BilimWeb: Bir Bilim Ziyafeti!