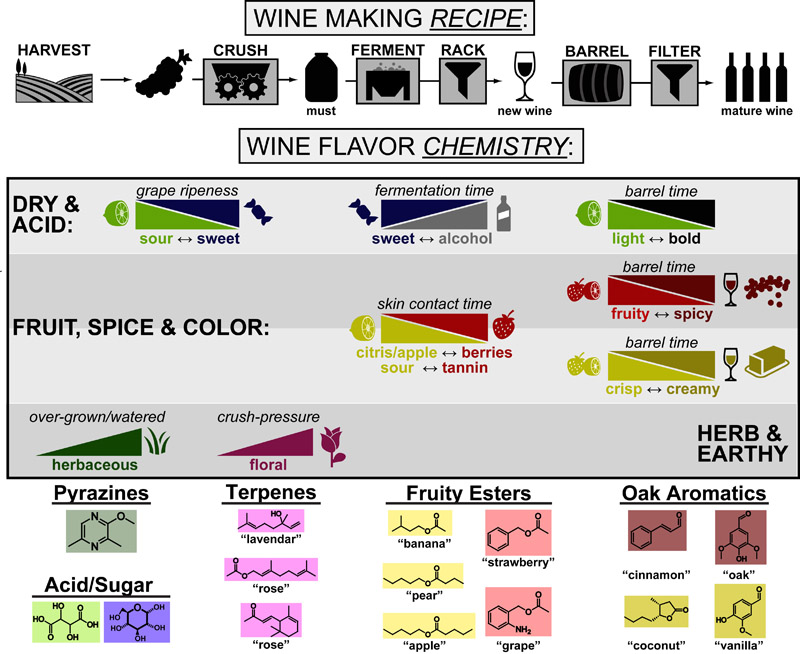
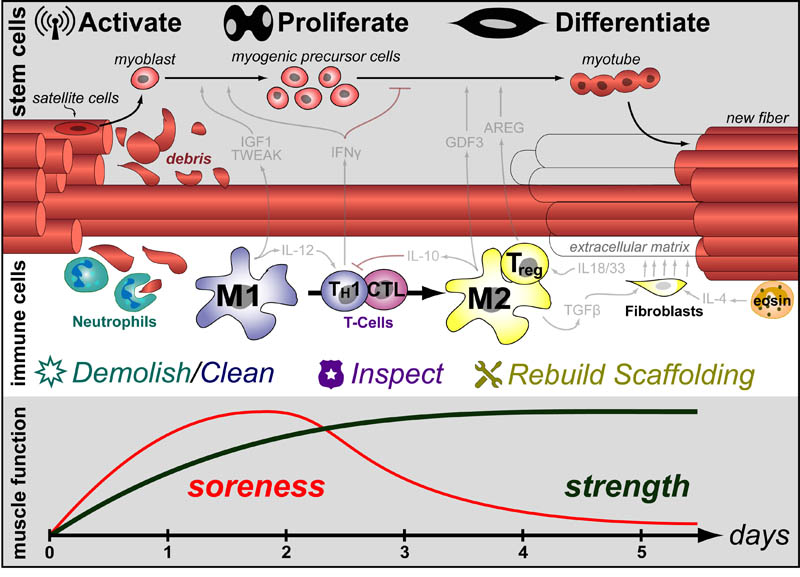
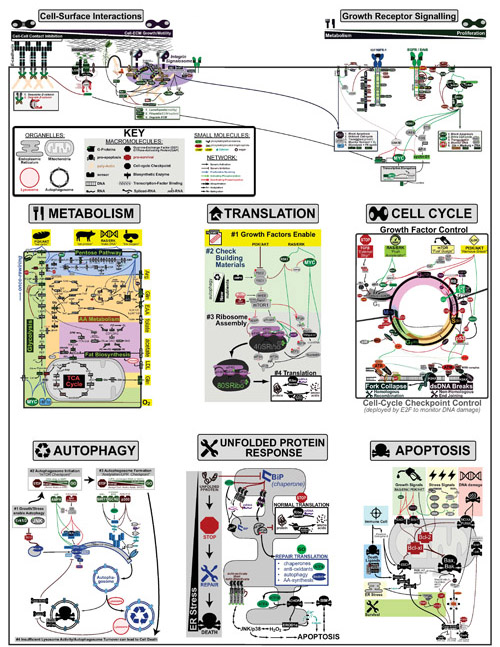
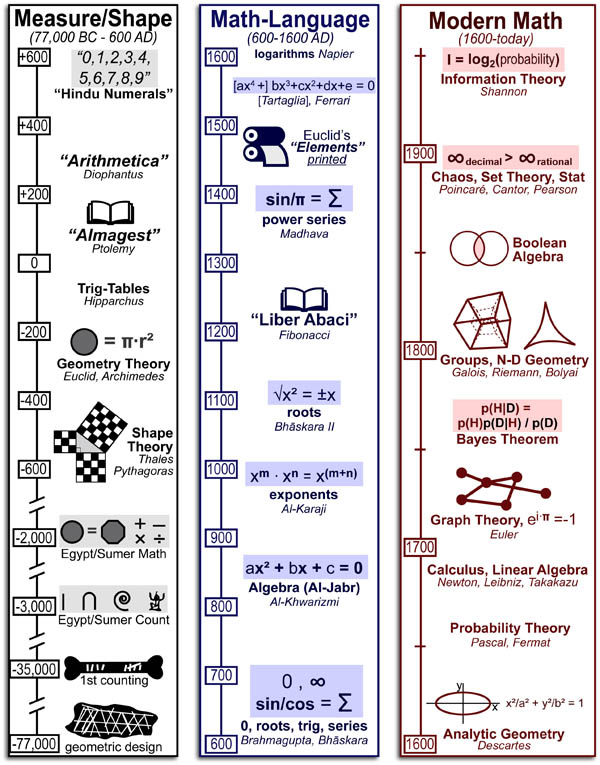
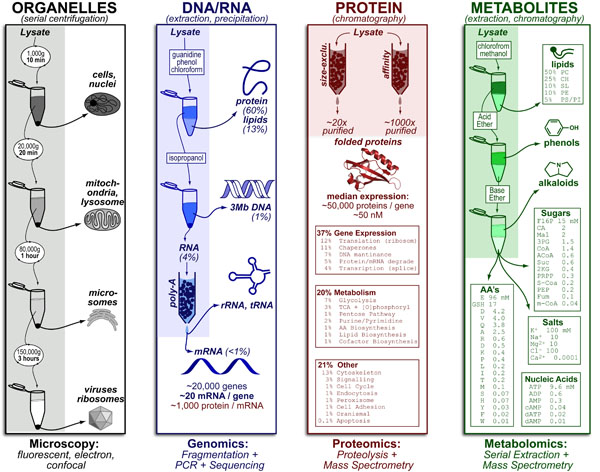
Extending our Wine Flavor Chemistry infographic to Whisky, we found there was alot of chemical overlap between flavor molecules found in whisky and the fruits and spices that whisky can taste like!
About Us
Practically Science was started by two Yale PhD students in 2012. Its goal is to make single-sheet summaries of common interdisciplinary methods, ideas, etc. Continue Reading →Please follow & like us :)
Blogroll
Site Map
-
Recent Posts